Essential software tools for the working scientist
The Scientist's Linux Toolbox

© Lead Image © KrishnaKumar Sivaraman, 123RF
Linux and science are a natural fit. These are a handful of essential software packages both for getting work done and presenting it to others.
Although Linux still occupies a small niche on the desktop among the population at large, it is much more popular among scientists from all disciplines.
It's tempting to say that's just because scientists are smart! But it's easy for me to understand Linux's appeal for scientists when I remember the problems caused by the use of proprietary operating systems (OSs and software in labs where I've worked).
For one particular piece of software, we had a site license that only permitted a certain number of people to use the program at a time. It would actually spy on the network and count up how many instances of the program were running. If I needed to run it and it refused, I had to run around the lab to find out if someone had just forgotten to exit the program.
People couldn't read each other's documents if they were made with the wrong version of Word. Programs would stop working after upgrading the OS, and, if they had been abandoned, you were out of luck without the source. Standard open source tools might not compile, because the OS vendor included outdated or even misnamed versions of standard libraries (Apple was notorious for this). Customizing the desktop was difficult and options were limited.
This is just the tip of the closed-source computing iceberg. When I was able to switch entirely to Linux, all these problems disappeared. I've been doing my work exclusively with Linux for years and could not imagine going back to the hostile world of proprietary software.
Another reason for the relative popularity of Linux among some scientists is that it is the OS of choice for such things as wiring together supercomputing clusters. There is a certain convenience in having a consistent environment shared between the remote compute resource and the box on your desk.
In the rest of this article, I survey some widely used free software. Except for some of the more specialized packages nearer the end of the article, I use all of this software myself and recommend it to the scientist switching to Linux who wants to get started with a set of powerful and reliable tools. (For those switching to Linux, see also the "Which Distribution?" box.)
Which Distribution?
All the software described here will work on any Linux distribution, so your best strategy is simply to use the distribution that is known to work well on your hardware. Since, as a scientist, you will probably wind up doing some serious calculations, you don't want to waste memory or cycles on inessentials. Because you will generally be working from the command line, regardless of your distribution, you should consider uninstalling or disabling any heavy desktop environment and replacing it with a lightweight window manager such as dwm [1].
Every piece of software I mention in this article is free and open source. All are available for Linux, most can also run on other free OSs, and some will even work on Apple and Windows machines.
Writing Papers
A scientist in any field will be writing papers, so this section is the most widely applicable. I recommend two nearly indispensable software packages.
The first is the TeX Live [2] distribution of LaTeX. This is a huge package that will install everything you need to typeset any kind of document, including a complete set of fonts, all the engines based on TeX (LuaTeX, pdfTeX, XeTeX, etc.), software for drawing diagrams, and much more. Do not install this from your distribution's package manager, because it will almost certainly be out of date.
It takes some time and effort to learn how to use TeX, but, especially if you are in a field where your papers will contain a lot of equations, it is really the only choice. Many journals accept TeX source, and some have their own templates that they require you to use. A convenient side effect is that you can use the same source to create a beautiful preprint.
Even if your papers never contain math, the LaTeX system is still a major convenience for the academic, because it handles references automatically, and it can generate bibliographies in any format.
The typical usage of the TeX system is to edit your source in the editor of your choice, embed the TeX markup, and process it through one of the engines (the modern choices are either LuaTeX or XeTeX) to create a PDF. However, you won't do it this way, because you will also install the second indispensable package: pandoc [3].
Pandoc is a "Swiss army knife" document conversion program. You write your papers in an extended version of Markdown, an intuitive markup that resembles the way people normally add emphasis and so on to text documents such as emails (i.e., *italic*). Pandoc can convert this markup to many formats, including HTML, as well as document formats such as ODT, DOCX, and TEX, which you can run through XeTeX or another TeX engine to produce a PDF. To include math or other elements that Markdown can't handle, just do it, and pandoc will know how to handle it. Pandoc is extremely useful for the working scientist, because some publications require a format other than TEX, and because it allows you to write one source for your paper and automatically create versions for preprints, the web, presentation slides, and more. Pandoc is also extensible [4] with user filters.
Figure 1 is a screenshot from my laptop showing this article as I write it in the Vim editor, on the left. On the right are three transformations of the article: as a PDF, processed through pandoc to XeTeX (top); the HTML version, directly from pandoc and rendered in a web browser (middle); and an ODT file, again directly from pandoc, viewed in LibreOffice (bottom). I'll include an equation to make it interesting (I am not including the part of the article with the command to include the screenshot, to avoid the creation of a spacetime singularity):
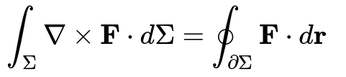
Making Graphs and Diagrams
There are many choices here; what you install and learn to use will be determined partly by your specialty. If you are a mathematician whose papers are full of things like commutative diagrams, you already have the best software for those purposes, because it comes with TeX Live: Learning how to use the TikZ drawing language, in which you can directly embed drawing instructions into the source for your paper, will be invaluable.
Many popular programming languages come with their own plotting systems, and using those may be a good choice if you are sure that you will always use the same language. However, if you want a portable solution that runs anywhere and is fast and stable when dealing with enormous datasets, consider gnuplot [5]. You will find a fairly recent version in your distribution's package manager, but for the very latest features, download and compile the source.
Gnuplot is an early open source program that predates Linux, but it is still actively developed, with new features [6] appearing regularly. It is the best choice for creating automated graphing pipelines that can work with simulations or data from any programming language or source. Gnuplot excels at automation because it is controlled through text scripts rather than with a GUI. It can create any type of output: PNG, SVG, dumb terminal, sixel, and many more, plus minimally interactive graphs for the web or using Qt, X11, and other GUI toolkits.
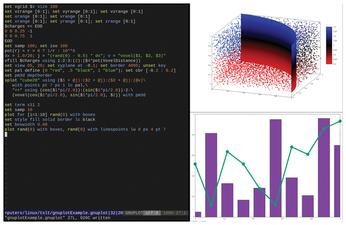
Gnuplot can create any type of visualization that a scientist might need, and the output is customizable to the last detail. Figure 2 is a screenshot from my laptop, showing a script in my editor on the left and the resulting graphs on the right.
Numerics
Linux has well-established, state-of-the-art compilers for C and Fortran: GCC and GFortran, respectively. The are both available in all package repositories. GCC is used to compile much system software, including the Linux kernel itself. GFortran is a capable compiler for Fortran simulation code and able to parallelize array expressions. For Fortran, there are many commercial compilers available as well and a surprising newcomer: the open source LFortran [7] compiler, which is based on LLVM and provides the user with an interactive REPL.
If you are involved with legacy simulation code, there is a good chance that you will find yourself using one of these tools. But if you are beginning a new project, my advice is to use neither C nor Fortran, but to head straight to the Julia website [8] and download this free, open source language for technical computing.
I wrote an article [9] about Julia about two years ago, introducing the syntax and use of the language. Since then, interest in this relatively new language has exploded among computational scientists in every field. This is due not only to its speed and ease of development, but to the ease [10] with which a scientist can mix and match different libraries to create new functionalities.
Up to here, I've treated subjects of general interest to most scientists. I'd like to turn now to a brief rundown of some software that is specific to several fields. There isn't space to survey the vast landscape of science, of course. But even if your field is not one of these few, this may give you an idea of the range and character of Linux tools focused on particular disciplines.
Buy this article as PDF
(incl. VAT)
